Deciphering the role of RNA editing in zebrafish development
- Principal Investigator: Dr Leszek Pryszcz, International Institute of Molecular and Cell Biology in Warsaw
- Project title: Deciphering the role of RNA editing in zebrafish development
- Funding scheme: POLONEZ 1, announced on 15 September 2015
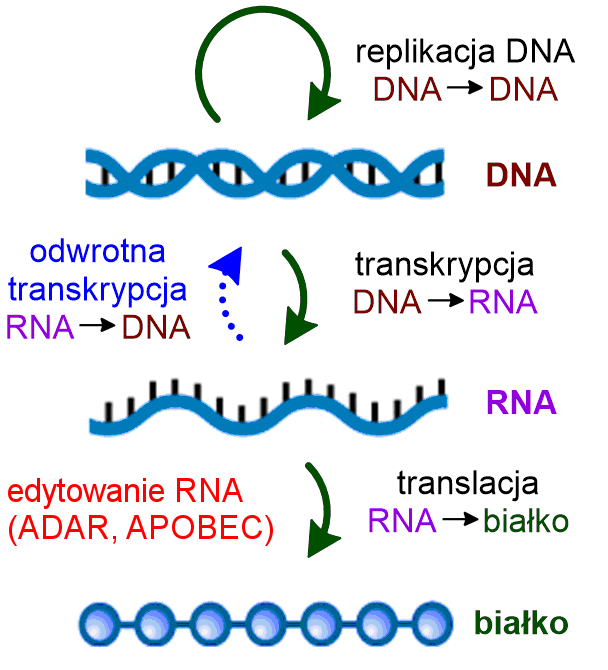
Central dogma of molecular biology and RNA editing. Source: Wikipedia, Creative Commons (author: Michał Sobkowski). Modified by Leszek Pryszcz.
RNA editing is a process whereby the RNA is modified after transcription. As such, it contradicts the central dogma of molecular biology, which holds that information flows from gene (DNA) through transcript (RNA) to protein (Fig. 1). Individual bases of the RNA may be inserted (added), deleted (removed) or converted into other bases through deamination, e.g. adenosine may change into inosine (A → I) or cytidine into uracil (C → U). In vertebrates, the process is catalyzed by enzymes from two different families: ADAR and APOBEC.
RNA editing was first described in the 1980s and its role has since been studied in many plant and animal species. It is known to affect both the physiology and the behaviour of various animals, ranging from insects to mammals. It is also a defence mechanism against RNA viruses and the expansion of mobile elements (i.e. Alu retrotransposons) present, among others, in the human genome.
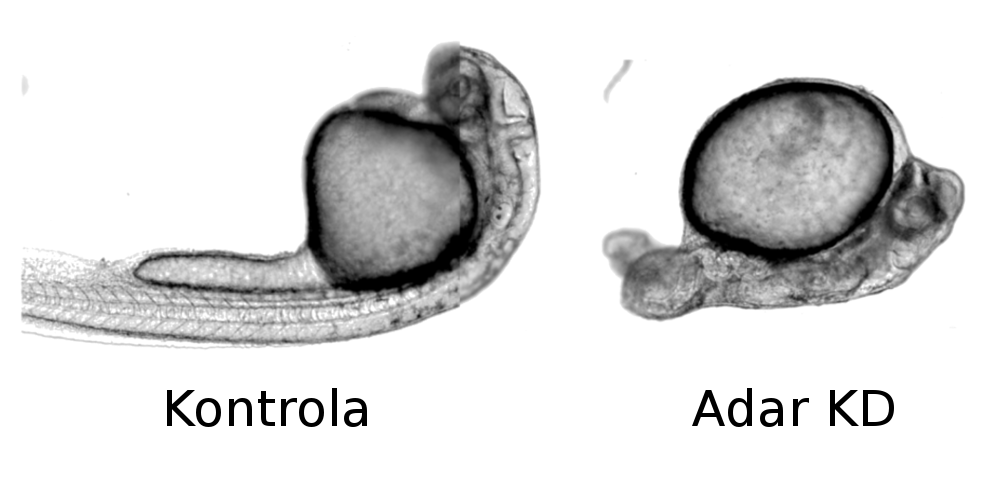
My project focuses on the role of RNA editing in early vertebrate embryonic development. The mechanism is studied on zebrafish (Danio rerio), one of the model species for research on vertebrate growth because of the easy maintenance of adult specimens and our ability to produce large quantities of zebrafish embryos. ADAR proteins are found in the ovum and persist throughout the embryonic development of the fish (Fig.2). Interestingly, their absence leads to significant abnormalities in early development (Fig.2). Changes can be observed as soon as three hours after fertilization; embryos deficient in the protein die on the very first day.
Edited RNA bases can be detected by comparing transcript sequences with the corresponding genes by means of next-generation sequencing (NGS). This method allowed us to identify transcripts edited by ADAR proteins. Many serve as key regulators of early development, responsible, for instance, for the normal definition of the anterior-posterior axis of the body. A detailed investigation into the role of RNA editing in early vertebrate development will contribute to a better understanding of gene expression regulation during embryonic growth.
I am working on my research at the Laboratory of Zebrafish Developmental Genomics, led by Dr Cecilia Winata at the International Institute of Molecular and Cell Biology in Warsaw. Apart from giving me access to cutting-edge technologies in molecular biology and promoting my personal growth, the POLONEZ programme has provided me with an opportunity to cooperate with researchers from all around the globe.
Dr Leszek Pryszcz
Science has fascinated Leszek Pryszcz ever since his early years. After completing his BSc in biotechnology at the University of Silesia in Katowice, he left for the UK, where he earned a MSc with honours in bioinformatics at the University of York in 2009. However, his research adventure only really began during his PhD studies. In 2009, he won a prestigious scholarship from La Caixa to study at the Universitat Pompeu Fabra in Barcelona. He conducted his PhD research project at the Comparative Genomics Lab under Dr Toni Gabaldón in the renowned Centre for Genomic Regulation (CRG). In 2015, he moved to the International Institute of Molecular and Cell Biology in Warsaw, where he has conducted his POLONEZ 1-funded research project since 2017.
Beyond his work in science, he is very involved in sharing skills and knowledge with the scientific community. He is the founder of #NGSchool, a non-profit organization, which has organized bioinformatics courses and summer schools since 2016.

Date of publication: 2nd Jan, 2018

